DNA REPLICATION
DNA REPLICATION INITIATION IN HUMANS
DNA replication is fundamental to life. This multistep process is highly dynamic and requires complex coordination for successful replication. The first step in DNA replication is the binding of a multi-subunit AAA+ ATPase called the Origin Recognition Complex (ORC1-6) to origins of replication on DNA. ORC plays a central role in origin selection, in which ORC binds to particular loci on chromosomes and recruits other replication factors to these replication start sites. In humans and higher eukaryotes, the mechanism of origin specification is unclear. Unlike yeast ORC, which binds to specific DNA sequences, human ORC has no apparent sequence specificity. Once ORC binds to DNA, the complex first recruits another AAA+ ATPase protein called CDC6, forming a complete ring that encircles dsDNA. This then allows ORC to recruit and subsequently load the hexameric AAA+ ATPase MCM helicase (MCM2-7) around DNA. The helicase then unwinds dsDNA and allows access to DNA polymerases for DNA synthesis to occur at replication forks.
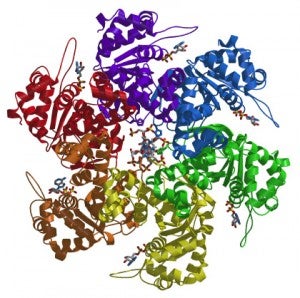
We are interested in unraveling how ORC orchestrates the events of DNA replication initiation. We have determined the first structure of human ORC with cryo-electron microscopy (cryoEM) and X-ray crystallography. Using advanced cryoEM techniques, we were able to determine the structure of the complex to higher resolution, and surprisingly uncovered five different states of the complex that revealed its dynamic nature (see figure). We determined that the flexibility is introduced by the binding of ORC1 to the ORC2-5 complex. The multiple states reveal two dynamic domains at the ring opening of the complex that are influenced by both nucleotide and DNA binding. In addition, we observed a hinge point at the interface between ORC3 and ORC5 that may serve an important role during binding to DNA and/or translocation along DNA.
We continue to investigate the upstream and downstream events in human DNA replication initiation and the roles of nucleotide binding and hydrolysis in influencing complex dynamics in these processes.
For more, please watch Matt Jaremko’s talk on ORC dynamics here.
THE DIFFERENT FACES OF E1 - A REPLICATIVE HEXAMERIC HELICASE
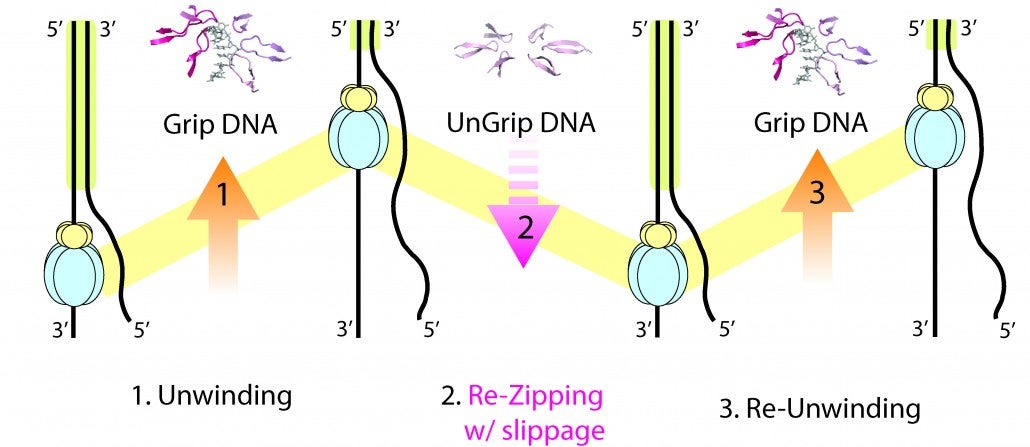
Strand separation required for DNA replication is mediated by a helicase enzyme, a molecular machine that uses the energy derived from ATP-hydrolysis while moving along the DNA. Our crystal structures of the dsDNA binding domain (DBD) of the replicative helicase E1 from papillomavirus in various stages of assembly, led us to propose a model in which the transition from the dimer to the ultimate double hexamer, results in strand separation. The loading and assembly of this protein separates the double helix, such that each hexameric helicase encircles one strand of DNA. Once assembled, the helicase uses its ATP-driven motor to translocate on the DNA or “pump” the ssDNA through the hexameric ring. Several competing mechanisms for helicase unwinding were proposed. Having determined a structure of hexameric E1 with ssDNA discretely bound in the central channel and nucleotides at the subunit interfaces, we showed that only one DNA strand passes through the hexamer channel and that the DNA-binding hairpins of each subunit form a spiral staircase that sequentially tracks the DNA backbone. The nucleotide configurations at the subunit interfaces indicate that each subunit sequentially progresses through ATP, ADP and apo states, while its associated DNA-binding hairpin travels from the top to the bottom of the staircase, each escorting one nucleotide of ssDNA through the channel, as if six hands grab the DNA and upon ATP hydrolysis and ADP release pull it through the channel. With this unique look into the mechanism of translocation of this molecular machine along DNA, we have focused on mechanistic aspects of the enzyme in solution.
By taking a multi-faceted approach including single-molecule and ensemble FRET (Förster Resonance Energy Transfer) methods, we have found that E1 is oriented with the N-terminal side of helicase facing the replication fork, consistent with the crystal structure. We also showed that E1 generates strikingly heterogeneous unwinding patterns stemming from varying degrees of repetitive movements that are modulated by the DNA-binding domain. Furthermore, our studies found that DNA-binding domain promotes the assembly of E1 helicase onto a forked DNA substrate, acting as an allosteric effector of the helicase. Our studies reveal previously unrecognized dynamic facets of replicative helicase unwinding mechanisms, adding another layer of complexity in the workings and regulation of DNA replication.